Susanna Röblitz appears in #UiBforklart
Professor and Group Leader at CBU Susanna Röblitz has made an appearance in #UiBforklart (“UiBexplained”). #UiBforklart is a series of short, outreach-oriented posts in social media, that aim to explain various topics and research areas in an easy-to-understand way. In her contribution to the series, Susanna shares insight into her research on Mathematical Systems Biology, and how the Trond Mohn Foundation has been critical for developing her particular field of research.
Click here to see the post on facebook, and click here to see the post on Linkedin.
Read more about CBU’s research on Mathematical Systems Biology.
March 11, 2025


Sushma Grellscheid appears in #UiBforklart
Professor and Group Leader at CBU, Sushma Grellscheid, is featured in #UiBforklart (“UiBexplained”) this week. #UiBforklart is a series of short, outreach-oriented social media posts that aim to explain various topics and research areas in an easy-to-understand way. In her contribution, Sushma shares insights on how cells are the fundamental units of life and how our understanding of their structure has evolved significantly through interdisciplinary research at CBU, particularly the study of droplets in cells and their role in understanding diseases like ALS. Support from the Trond Mohn Foundation has been crucial in advancing Sushma’s and her team’s work in Bergen.
Click here to see the post on facebook, and click here to see the post on Linkedin.
Read more about the Grellscheid group’s research here.
February 10, 2025
Håkon Dahle appears in #UiBforklart
Professor and Group Leader at CBU Håkon Dahle is making an appearance in #UiBforklart (“UiBexplained”) this week. #UiBforklart is a series of short, outreach-oriented posts in social media, that aim to explain various topics and research areas in an easy-to-understand way. In his contribution to the series, Håkon shares insight into his research on microbial systems, and how the Trond Mohn Foundation has been critical for developing his particular field of research.
Click here to see the post on facebook, and click here to see the post on Linkedin.
Read more about CBU’s research on Marine systems biology.
January 20, 2025


Markus Miettinen appears in #UiBforklart
Associate Professor and Group Leader at CBU Markus Miettinen featured in #UiBforklart (“UiBexplained”) last month. #UiBforklart is a series of short, social media posts aimed at explaining various topics and research areas in an easy-to-understand way. In his contribution, Markus explains how his research utilizes advanced simulations to improve treatment and diagnosis of Huntington’s disease, and how the Trond Mohn Foundation enables his group to undertake research projects they otherwise would not have the capacity to do.
Click here to see the post on facebook, and click here to see the post on Linkedin.
Read more about CBU’s research on High-fidelity Biomolecular Modelling.
January 8, 2025
Anagha Joshi appears in #UiBforklart
Professor and Group Leader at CBU Anagha Joshi is making an appearance in #UiBforklart (“UiBexplained”) this week. #UiBforklart is a series of short, outreach-oriented posts in social media, that aim to explain various topics and research areas in an easy-to-understand way. In her contribution to the series, Anagha shares her journey into and motivation for researching women’s health, and how the Trond Mohn Foundation allows her to leverage her research to shape future public health policy.
Click here to see the post on facebook, and click here to see the post on Linkedin.
Read more about CBU’s research on women’s health.
December 17, 2024

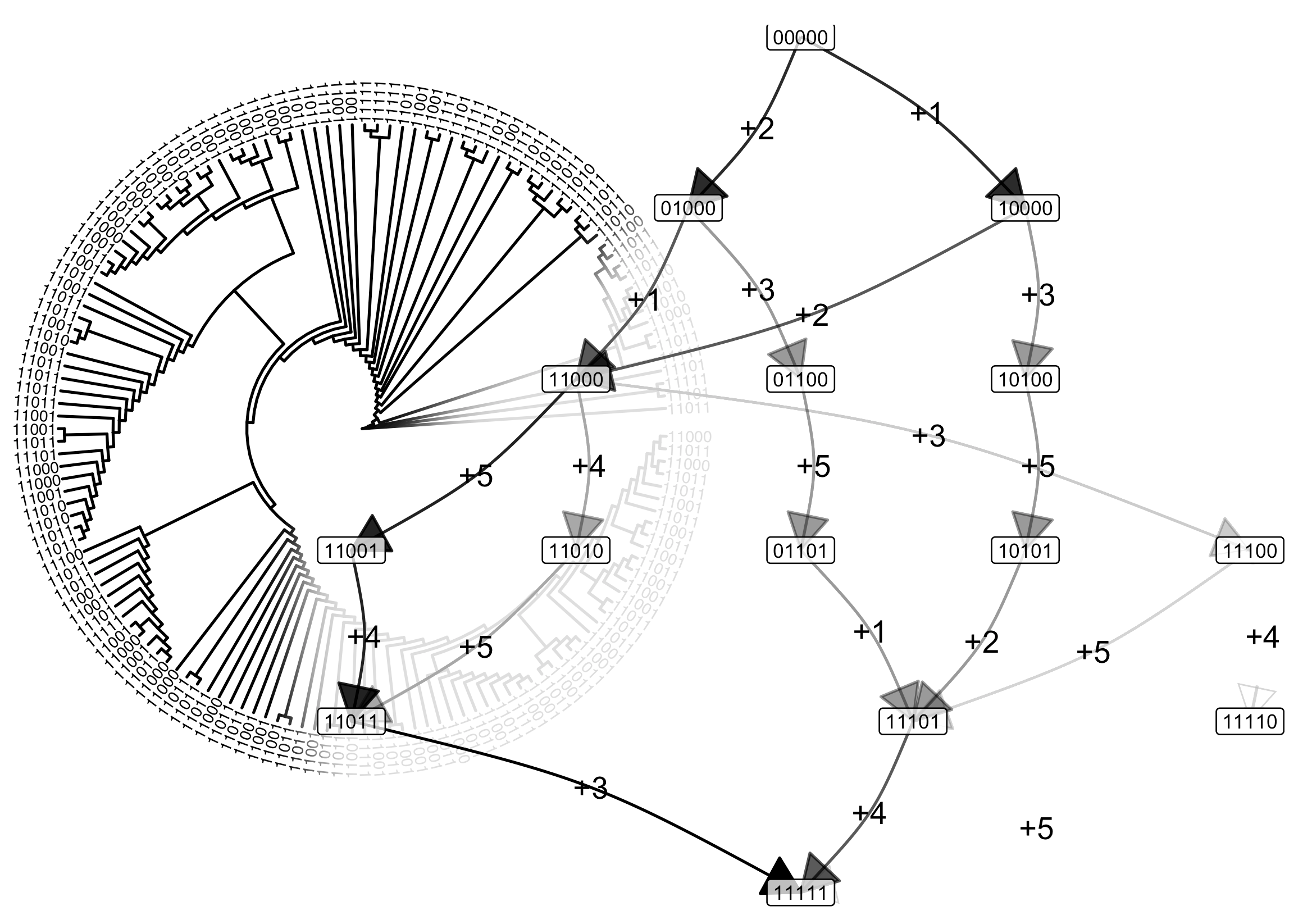
Learning how reversible processes evolve
When tumours evolve inside a patient, they acquire more and more mutations in their DNA. Lots of computational approaches try to learn how this sort of “accumulation” evolution happens – in cancer and many other systems. But many of those approaches don’t account for the fact that mutations (or other features) can be lost as well as gained. A new paper in Bioinformatics borrows from phylogenetic methods that can account for gains, losses, and arbitrary influences between features.
December 12, 2024
CBUs Franziska Görtler receives high-ranking grant
Researcher and member of CBU Franziska Görtler has been awarded funding from RCN (Research Council of Norway) FRIPRO for early career researchers. With this she will receive 8 million NOK to establish and conduct research that will, over three years from 2026, work on groundbreaking projects with significant medical benefits.
In addition to being a valuable member of CBU, Franziska is a researcher at the Department of Clinical Science at the Faculty of Medicine. Her research mainly focuses on algorithm development for immune cell deconvolution in tissues.
Through loss-function learning optimziation, she wants to find out more about the dissection of the cellular distributions and the cell-type specific regulation patterns in spatial transcriptomics data.
Big congratulations from all of us!
Here is a link to the full list of the grant recipients.
December 10, 2024


Top European grant to CBUs Marc Vaudel
Associate Professor and member of CBU Marc Vaudel has been awarded a prestigious ERC Consolidator Grant. With this he will receive around 23 million NOK to establish a research group that will, over the next five years, work on groundbreaking projects with significant societal benefits.
In addition to being a valuable member of CBU, Marc is a researcher at the Department of Clinical Science at the Faculty of Medicine. His research mainly focuses on women’s pregnancies, symptoms and complications during pregnancy, as well as what controls both the mother’s and the fetus’s genetic material.
Through advanced computer modeling, he wants to find out more about women’s symptoms early in pregnancy. The symptoms can provide important information about the mother’s risk of cardiovascular disease later in life.
Big congratulations from all of us!
December 4, 2024
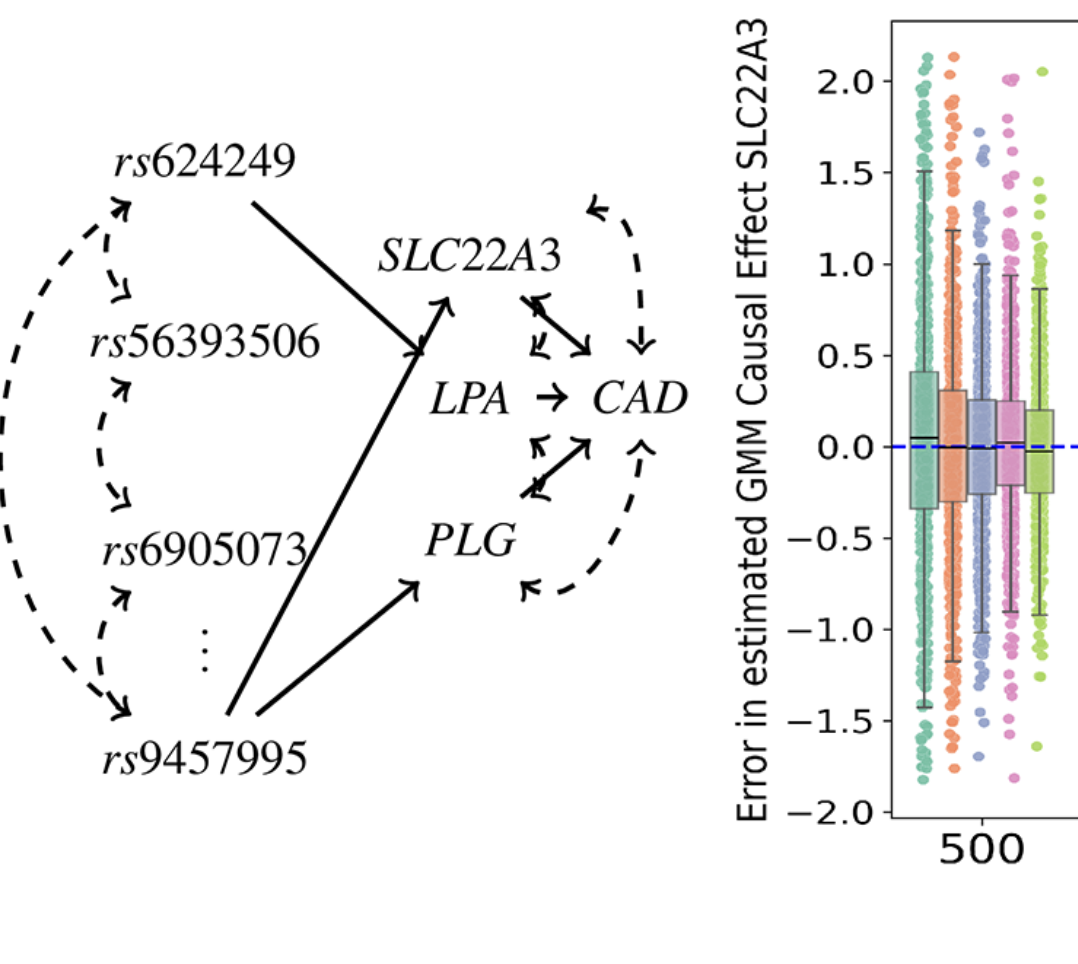
Predicting disease genes with causal inference
Scientists are trying to figure out which genes cause diseases like heart disease and which ones don’t. This is tricky because many genes are close together on the genome and are affected by the same DNA variations. Mariyam Khan, a PhD student in the Michoel group, used an old mathematical idea from the theory of causal inference to improve a method called Mendelian randomization. This method helps scientists understand how certain factors affect health. Her new approach (in PLoS Genetics) can identify which genes are likely causing diseases, even in complicated areas of the genome. When tested on data from 600 heart surgery patients, it confirmed that some genes, like PHACTR1 and ADAMTS7, are involved in heart disease and found new genes that need more research.
November 11, 2024
Postdoc Sean Alexander Bankier – Young Scientist Award 2024
Japan Society for Human Genetics has awarded Sean Bankier, postdoctoral fellow in the Michoel Group, with the Young Scientist Award 2024. JHG Young Scientist Award recognises articles by young researchers that have made significant contribution to Journal of Human Genetics, thanks to their scientific excellence and impact in the field of human genetics.
Bankier received the award for the article “Variation in the SERPINA6/SERPINA1 locus alters morning plasma cortisol, hepatic corticosteroid binding globulin expression, gene expression in peripheral tissues, and risk of cardiovascular disease“
November 4, 2024
Learning evolutionary and progression pathways over time
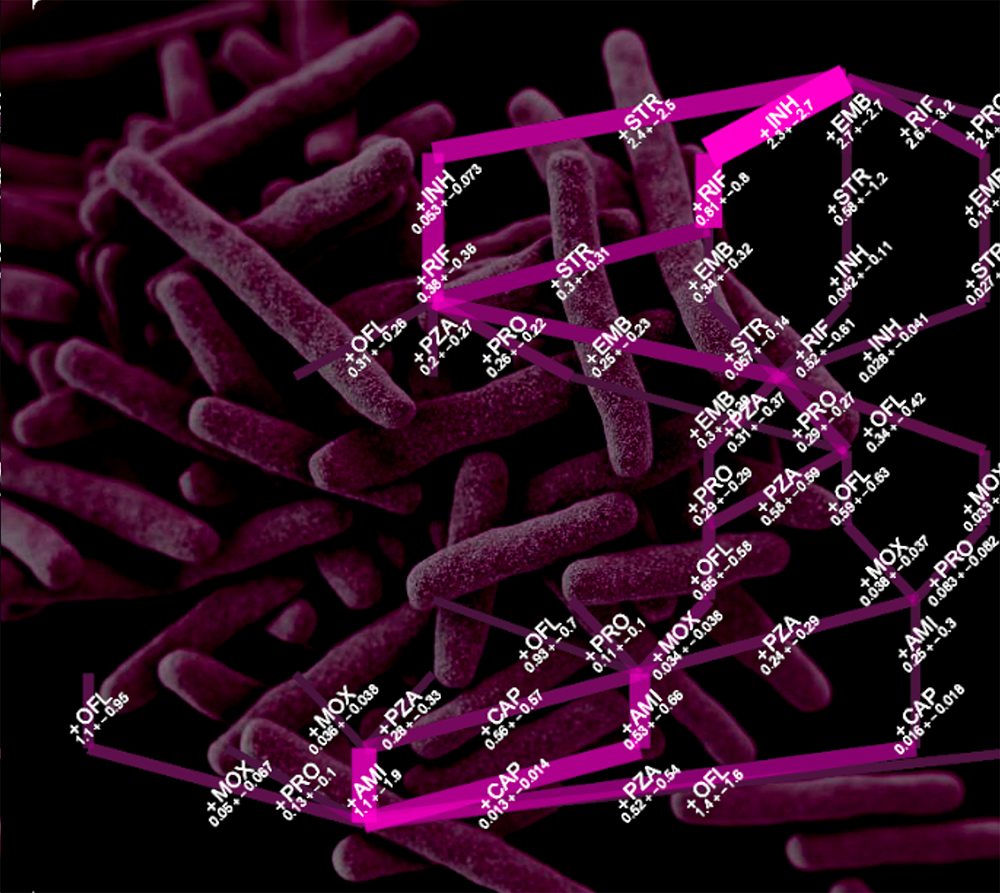
Many important processes in biology and medicine involve a progressive buildup of features over time. These might be, for example, the accumulation of different mutations as cancer progresses, or the evolution of bacteria to be resistant to more and more drugs. In PLoS Computational Biology, we have developed a new algorithm called HyperTraPS-CT uses data to learn the details of how these features build up over time. We demonstrate its use in learning about leukemia progression and tuberculosis drug resistance.
September 4, 2024
Lipid extraction by the STARD2 lipid transfer protein
Reza Talandashti, PhD candidate in the Reuter group, used molecular dynamics simulations and free energy calculations and proposes a model for the mechanism by which STARD2 extracts choline lipids from the endoplasmic reticulum membrane. Reza’s work is published in the Journal of Physical Chemistry Letters.
August 6, 2024
Feast or famine: how sea anemones resize their bodies to survive
Unlike other animals, sea anemones grow and shrink dramatically when fed or starved. Their bodies can get over ten times bigger when they consume a lot of food, then shrink back ten times smaller again. A collaboration between the Computational Biology Unit and the Michael Sars Centre, published in Development, has revealed how these creatures manage these enormous changes in body size.
You can also read the UiB news story.
July 9, 2024
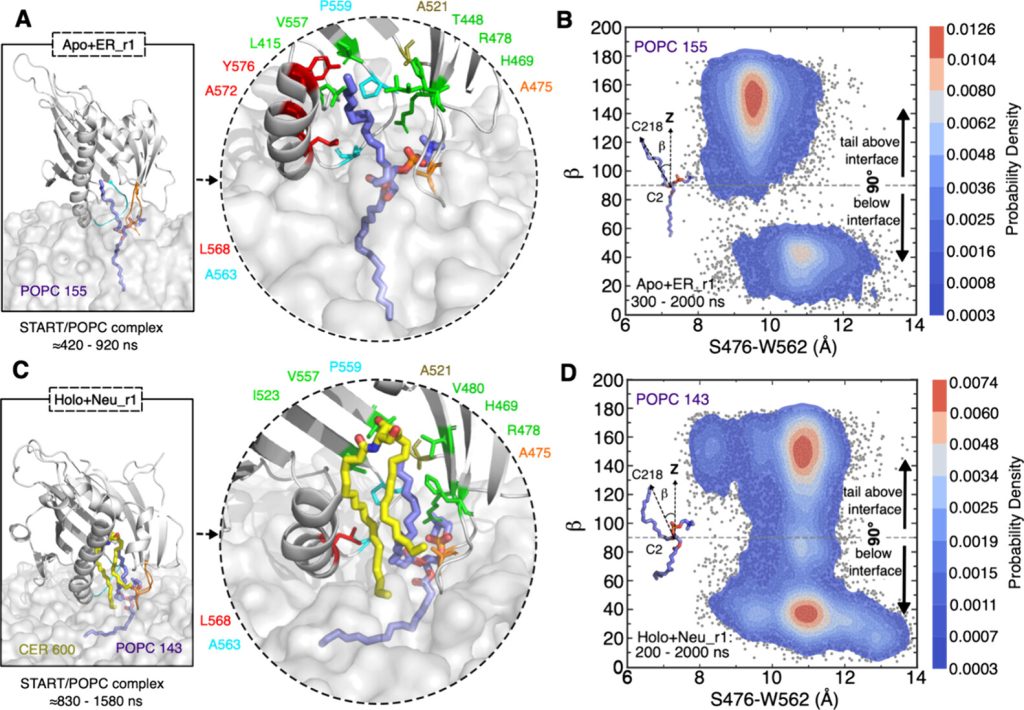
Molecular simulations shed light on lipid transfer mechanism
Lipid transfer proteins (also known as LTPs) facilitate non-vesicular lipid transport between organelle membranes and regulate various cellular processes, including signal transduction and lipid metabolism. The mechanisms by which LTPs bind to membranes and extract/release lipids from/into well-packed membranes are extremely complex. They are also extremely challenging to address with experimental set-ups. In two recently published works, we used molecular dynamics simulations to propose mechanistic models for two LTPs transferring either cholesterol (STARD 4), or ceramide (CERT/STARD11). Both models could be validated by experiments and reveal conformational changes undergone by the proteins in the process, and the active role played by the membrane in the lipid extraction and release processes. The works are published in J.Mol.Biol (doi.org/10.1016/j.jmb.2024.168572) and J.Phys.Chem.B (doi.org/10.1021/acs.jpcb.4c02398).
June 22, 2024
Universal evolution in mitochondria and chloroplasts
Mitochondria and chloroplasts power complex life. They have their own genomes, which encode vital parts of bioenergetic machinery. These genomes have become reduced over evolutionary history. Previous research from the CBU has hypothesised “universal” rules shaping these genomes. In Molecular Biology and Evolution, our new research supports this hypothesis, by showing that evolutionary reduction has occurred similarly in mitochondria and chloroplasts.
May 17, 2024
Plants’ ingenious defense against mutational damage
Mutation damages vital mitochondrial DNA and chloroplast DNA inside plant cells. New research from the CBU, with collaborators from Colorado, has shown how plants use randomness to deal with this damage. Published in New Phytologist (and featured on the cover as in the picture; you can watch a Youtube video summary and read the UiB news story), the team used mathematical models and new experiments to show how plants randomly “sort out” DNA as they develop. This ensures that some offspring of a damaged plant inherit intact DNA, preventing the full amount of damage being inherited from mother to offspring.
November 5, 2023
Postdoc Sean Alexander Bankier – new manuscript
Genome-wide association meta-analysis (GWAMA) by the Cortisol Network (CORNET) consortium identified genetic variants spanning the SERPINA6/SERPINA1 locus on chromosome 14 associated with morning plasma cortisol, cardiovascular disease (CVD), and SERPINA6 mRNA expression encoding corticosteroid-binding globulin (CBG) in the liver. These and other findings indicate that higher plasma cortisol levels are causally associated with CVD; however, the mechanisms by which variations in CBG lead to CVD are undetermined. Using genomic and transcriptomic data from …
September 8, 2023
Socialising in the “flatland” of the plant cell
Chloroplasts catch light to power plants. Under stress, they form mysterious structures called “stromules”. Stromules are long, thin bits of membrane that stretch away from the chloroplast, and it’s not clear what they’re for. A new paper from a CBU-Halle collaboration (featured as a highlight) in Plant and Cell Physiology explores these structures. It shows that in many plant cells, the environment is very thin (less than a micron wide). Computational modelling shows that stromule formation is an efficient way for chloroplasts to interact with other parts of the cell in this remarkable “flatland”.
September 2, 2023
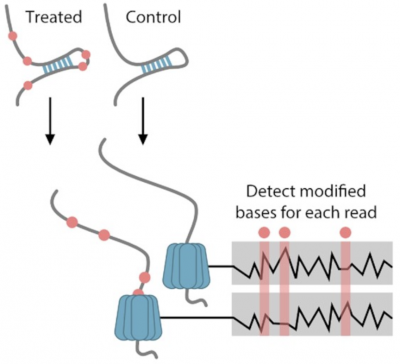
Probing the structure of individual RNA molecules
RNA molecules can form secondary and tertiary structures that can regulate their localization and function. These structures are rarely static and different copies of the same RNA can often adopt many different conformations. Despite this most methods are only capable of characterizing RNA at the consensus level, looking at the average over RNA molecules.
A new paper in Nucleic Acids Research from Teshome Bizuyaheu in the Valen group addresses this by developing a method to probe for structure …
October 28, 2022
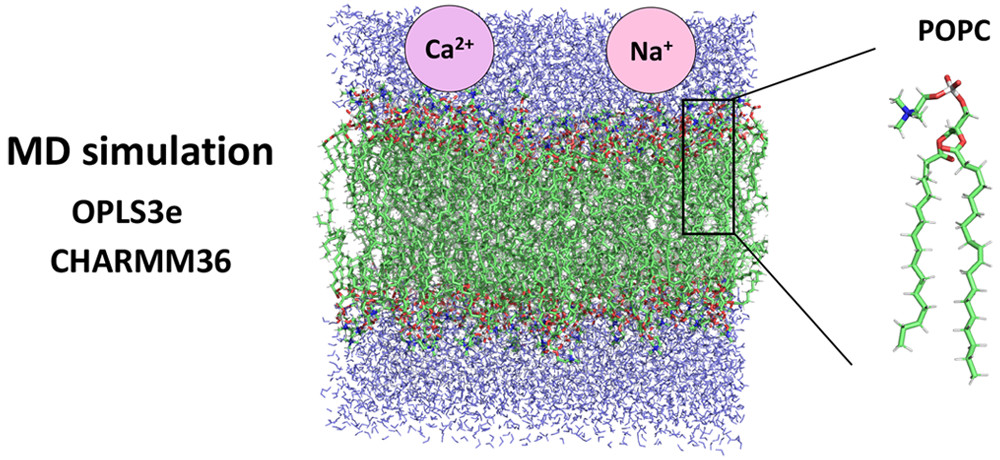
Structure of POPC Lipid Bilayers in OPLS3e Force Field
It is crucial for molecular dynamics simulations of biomembranes that the force field parameters give a realistic model of the membrane behavior. In this study, we examined the OPLS3e force field for the carbon–hydrogen order parameters SCH of POPC (1-palmitoyl-2-oleoylphosphatidylcholine) lipid bilayers at varying hydration conditions and ion concentrations. The results show that OPLS3e behaves similarly to the CHARMM36 force field and relatively accurately follows the experimentally measured SCH for the lipid headgroup, the glycerol backbone, and the acyl tails. Thus, OPLS3e is a …
September 8, 2022