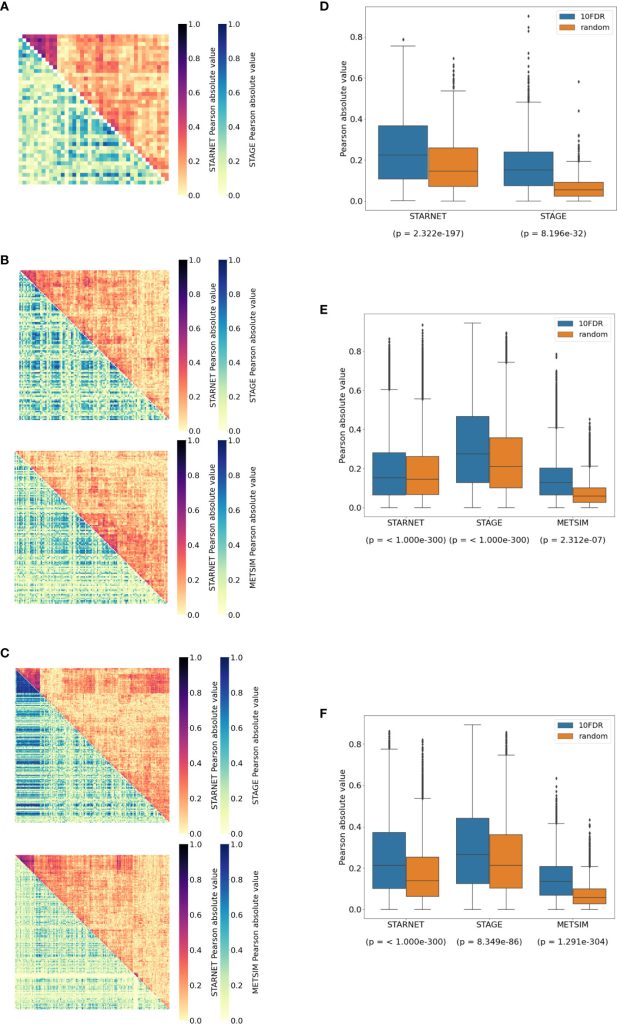
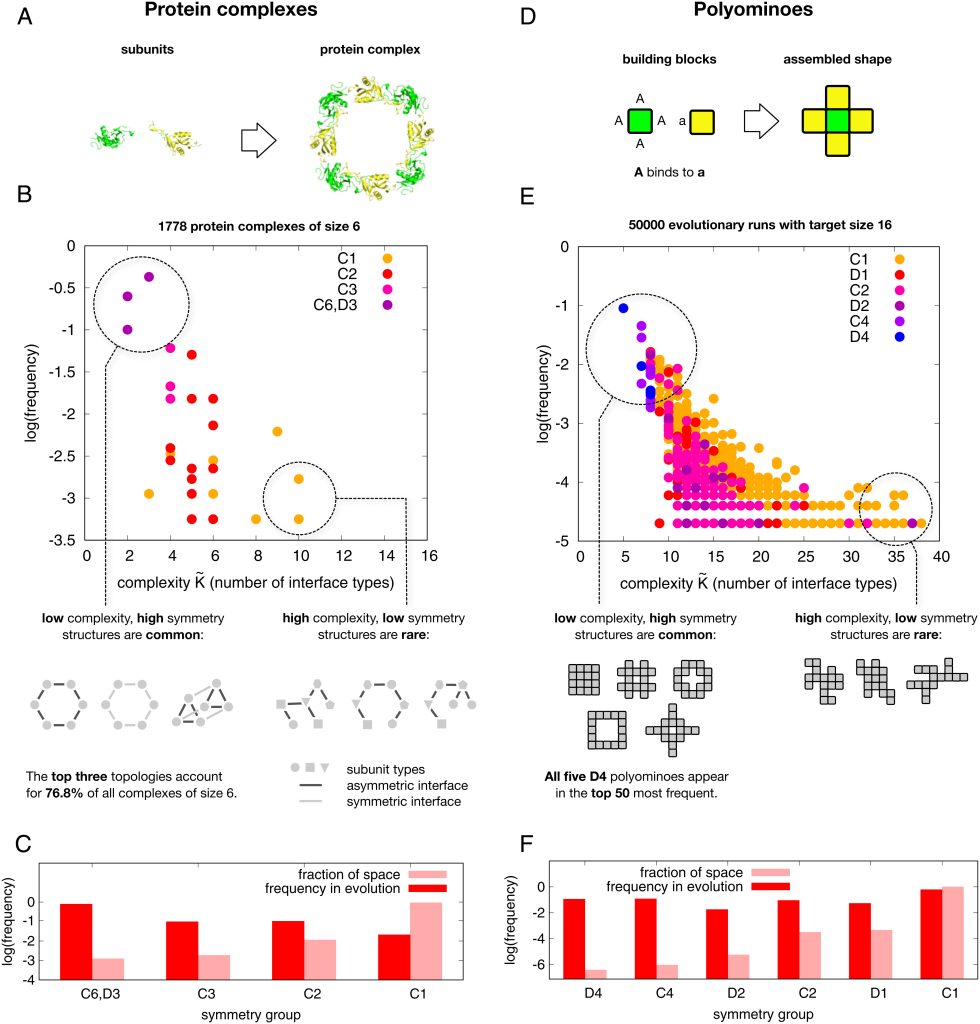
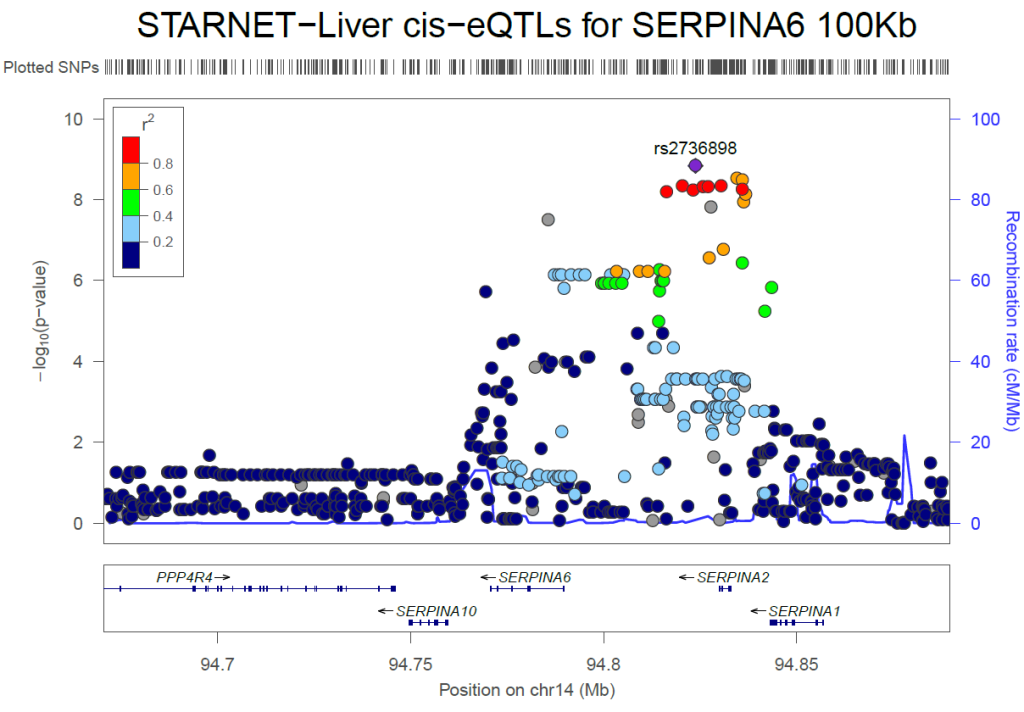
We are please to announce that our postdoc Sean Alexander Bankier together with the co-authors published the manuscript: “Plasma cortisol-linked gene networks in hepatic and adipose tissues implicate corticosteroid-binding globulin in modulating tissue glucocorticoid action and cardiovascular risk”. Frontiers | Plasma cortisol-linked gene networks in hepatic and adipose tissues implicate corticosteroid-binding globulin in modulating tissue …