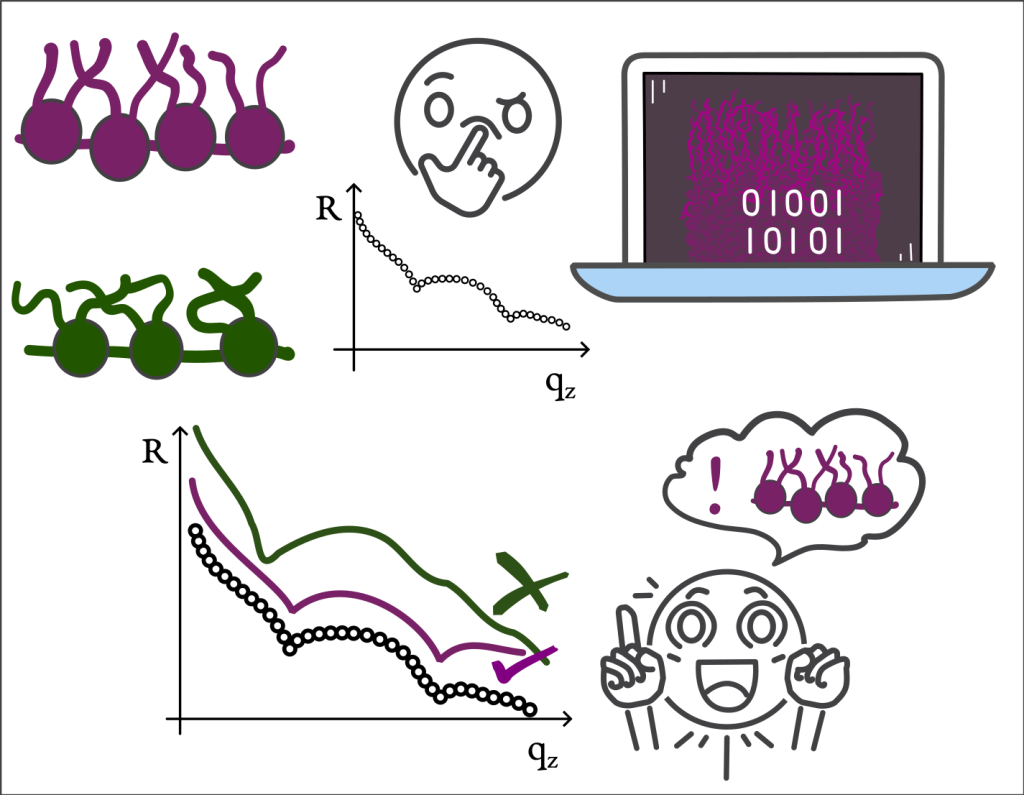
In the first paper, published in Colloids and Surfaces A: Physicochemical and Engineering Aspects, Markus and his team propose a method to correct inconsistencies in lipid monolayer experiments. They combine X-ray reflectometry with molecular simulations to determine accurate molecular areas. This correction improves the reliability of surface-pressure–area isotherms. It was successfully applied to several lipid types, enhancing reproducibility.
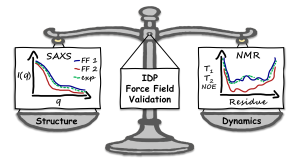
The other paper, published in Journal of Chemical Theory and Computation, evaluates 20 molecular dynamics (MD) force fields for their ability to simulate the structure and dynamics of COR15A, an intrinsically disordered protein (IDP). Using X-ray scattering and NMR data, the researchers identify DES-amber as the most accurate force field, capturing both structural and dynamic features of COR15A. The study highlights that water models tailored for IDPs improve simulation accuracy more than reducing protein–protein interactions does. This work emphasizes the importance of validating MD models against both structural and dynamic experimental data for IDPs.