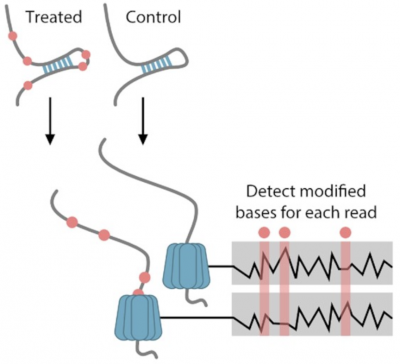
RNA molecules can form secondary and tertiary structures that can regulate their localization and function. These structures are rarely static and different copies of the same RNA can often adopt many different conformations. Despite this most methods are only capable of characterizing RNA at the consensus level, looking at the average over RNA molecules. A new …