Our PhD student Muhammad Ammar Malik and Professor Tom Luk Michoel published a paper in which they analyse the mathematical structure of a class of statistical models for learning hidden factors influencing gene expression data and show that a new algorithm based on the analytical results is orders of magnitude faster than the standard algorithms …
Month: December 2021
A Method To Learn Hidden Factors Influencing Gene Expression Data
Muhammad Ammar Malik and Tom Luk Michoel published a paper in which they analyse the mathematical structure of a class of statistical models for learning hidden factors influencing gene expression data and show that a new algorithm based on the analytical results is orders of magnitude faster than the standard algorithms for solving this class of models. Read …
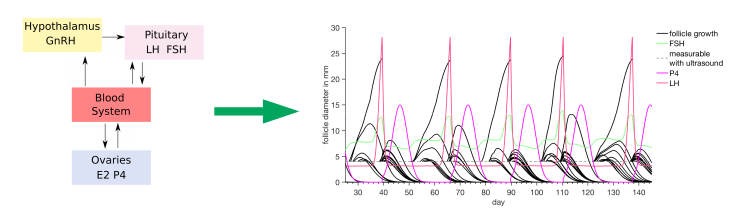
Mathematical Modeling and Simulation Provides Evidence for New Strategies of Ovarian Stimulation
The Röblitz group underpinned the theory that ovarian follicles grow in cohorts (“waves”) and added further scientific evidence about the efficiency of variable-start ovarian stimulation protocols by using mathematical modelling and numerical simulations. Read the paper here
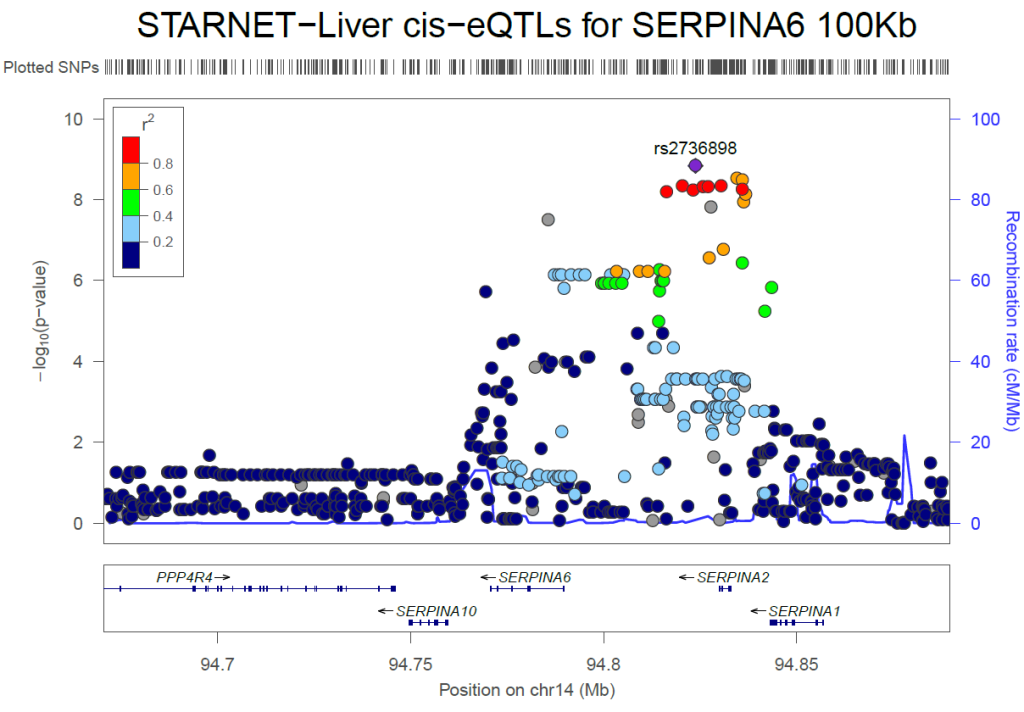
Genome-Wide Association Study For Plasma Cortisol
The stress hormone cortisol modulates fuel metabolism, cardiovascular homoeostasis, mood, inflammation, and cognition. Sean and Tom contributed to a genome-wide meta-analysis that mapped genetic variants associated with plasma cortisol levels and how they affect gene expression in adipose tissues. Read the paper here.
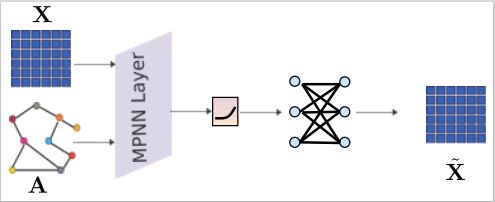
Graph neural networks for analysing biological networks
Ramin published a new Graph Feature Auto-Encoder for integrating biological networks with gene expression data which can impute missing values in for instance single-cell RNAseq data more accurately than existing methods. Read the paper here.
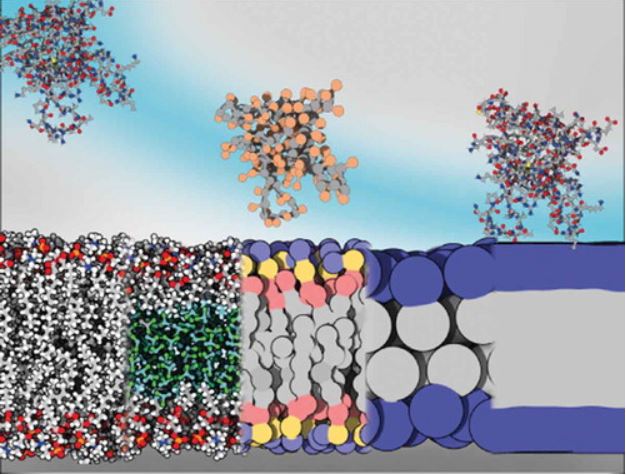
Membrane Models For Molecular Simulations Of Peripheral Membrane Proteins
The Reuter group wrote a short review on membrane models for simulations of peripheral membrane proteins (PMPs): which one to choose, when and why? Hopefully useful for getting into simulations of PMPs.
Martini 3: A General Purpose Force Field For Coarse-Grained Molecular Dynamics
We (the Reuter Group) are glad that we could contribute to the new version of the Martini coarse grained force field (Martini 3).
