We conduct computational and experimental studies of the membrane binding of peripheral membrane proteins. Experiments are performed in the group or conducted by collaborators. In particular, we have earned insights into the binding of serine protease 3 (PR3), the human N-acetyl transferase Naa60 and a bacterial Phospholipase C. We are also working on pattern identifications from large-scale sequence and structural data sets using bioinformatics analysis.
Peripheral membrane proteins bind temporarily to biological membranes. Unlike integral membrane proteins, the portion of the protein inserted into the lipid-bilayer is restricted to the interfacial area. In common with integral membrane proteins they share a paucity of structural data. Hence, we perform sequence and structural data analysis on peripheral proteins using bioinformatics approaches. One such outcome is our recent work highlighting hydrophobic protrusions model (Fuglebakk and Reuter, PLoS Comp. Bio., 2018).
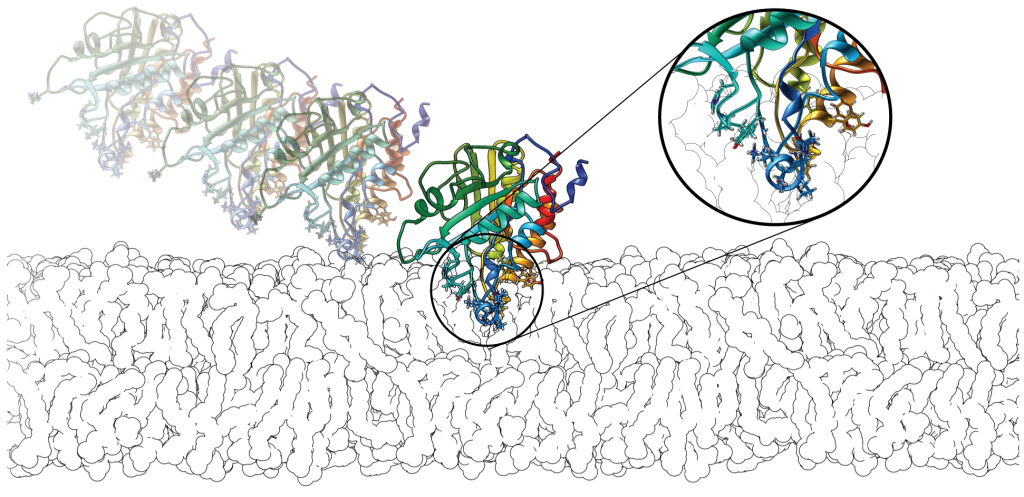
We are interested in understanding the peripheral membrane binding mechanisms and the role of different amino acid types in peripheral binding. Our ongoing efforts are particularly focused on the role of aromatic amino acids and their energetic contributions in the membrane binding using free energy calculations.
Based on our investigation on a bacterial phospholipase from Bacillus thuringiensis (BtPI-PLC), we suggested that choline-tyrosine cation-pi interactions at the membrane interface might be a novel anchoring mechanism for peripheral proteins (Grauffel et al., JACS, 2013). We evaluated the ability of the CHARMM force field to model pi-cation interactions between methylated ammonium and the three aromatic amino acids; we proposed a modification of the Lennard-Jones terms in CHARMM to best reproduce QM reference structures and potential energy surfaces (Khan et al., JCTC, 2016 & Khan et al., JCTC, 2019). We also propose a modification of the MARTINI2.2P force field (coined MARTINI2.3P, Khan et al., JCTC, 2020). We calculated that interfacial aromatics mediating cation−π interactions with choline-containing lipids can contribute as much to peripheral protein affinity for membranes as aromatics inserted below the phosphates of the lipid bilayer (Waheed et al, J Phys Chem Lett, 2019).
We could show that BtPI-PLC, unlike prototypical peripheral proteins, uses weak electrostatic interactions to bind to lipid vesicles (Khan et al., Biophys. J., 2016). A quantitative model for BtPI-PLC cell membrane interactions has been proposed by the group of our collaborator Anne Gershenson (UMass-Amherts) based on single molecule experiments and computations (Yang et al., JACS, 2015). Using a combination of in vitro and in silico mutagenesis conducted by the group of Anne Gershenson, our teams had earlier demonstrated how correlated motion in a bacterial phospholipases is important for their enzymatic activity (Cheng et al., Biophys. J., 2013). Recently we summarized these findings on BtPIPLC and other homologues with our collaborators in a review article (Roberts et al., Chem. Rev., 2018), where we also discussed different experimental and computational techniques (and their combinations) to study these types of peripheral proteins.
Together with the Arnesen’s group we have shown that the N-alpha acetyltransferse (Naa60) anchors to the Golgi membrane using two amphipathic helices located at its C-terminus (Aksnes et al., J. Biol. Chem., 2017). The Arnesen’s group had earlier revealed that Naa60 shows a distinctive localization to the secretory pathways and preferentially acetylates transmembrane proteins with an N-terminus facing the cytosol. We are further investigating the membrane binding orientation of the full structure of Naa60 which can elaborate the role of Naa60 in N-terminal acetylation of a transmembrane protein.
Our investigation on serine protease (PR3) with implicit membrane model reveals that PR3 is able to bind both anionic and neutral membranes but with a preference for negatively charged lipids (Hajjar et al., Proteins, 2008). We identified a unique membrane-binding site, which contains hydrophobic patch, carried by surface loops that insert into membrane. Simulations with explicit (all-atoms) bilayer models reveal the importance of interactions between aromatic amino acids and the cationic lipid headgroups (cation-pi interactions) (Broemstrup, PCCP, 2010). Our work (experiments and simulations) reveals PR3 has higher affinity towards zwitterionic membranes compared to its homologue HNE due to partitioning of aromatic amino acids in the membrane (Schillinger et al., BBA-Biomembranes, 2014).
Collaborators:
Work on serine proteases are done in collaboration with Themis Lazaridis (CCNY, USA), Véronique Witko-Sarsat (INSERM, France) and Øyvind Halskau (UiB, Norway).
Experimental work for bacterial phospholipases projects are done by our external collaborators- Anne Gershenson (UMassAmherst, USA) and Mary F. Roberts (Boston College, USA).
The N-alpha acetyltransferase is studied in collaboration with Thomas Arnesen’s research group (UiB, Norway).
Techniques:
Implicit membrane model (IMM1-GC), Continuum electrostatics, All-atom Molecular Dynamics (MD) and Coarse-Grained molecular dynamics (CGMD), Free energy calculations, Statistical analysis of protein structure databases, Surface Plasmon Resonance (SPR) Spectroscopy.
