Proteins’ functions are dependent on their structure and dynamics, two properties that do not easily let themselves observe directly from the lab bench. Powerful and enlightening alternatives are the methods of computational molecular modelling and simulations. Combined with protein bioinformatics to, for example, evaluate how general our findings are.
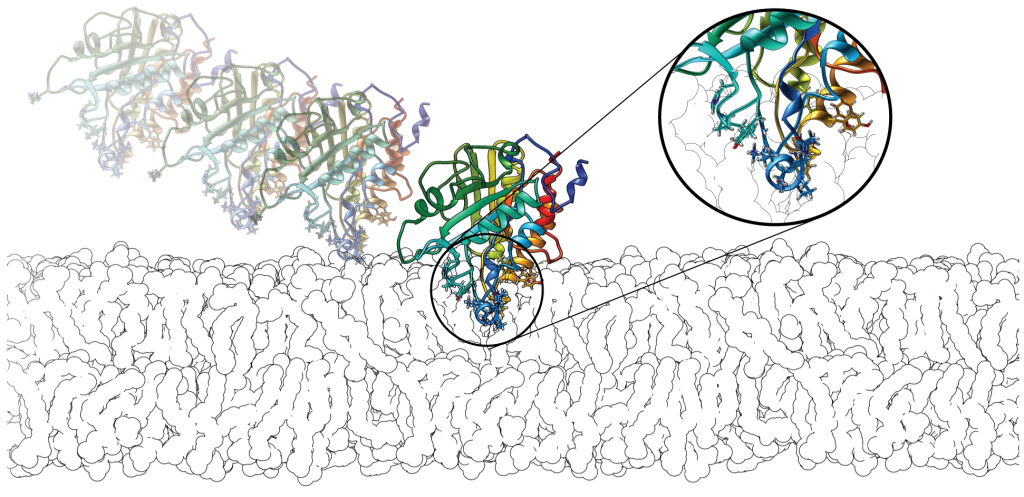
We look at the structure-dynamics-function relationships, its conservation in families and its importance for membrane peripheral binding and enzyme activity, including inhibitor and drug design.
We develop web tools such as WEBnma, for ENM-NMA, and PePrMInt, our new resource for exploration of peripheral membrane proteins surfaces, and of their hydrophobic protrusions.
